Background

What is a HMA?
Hydroxy-mandelic acid (HMA) is widely used in the production of aromatic drugs and spices[1]. It can be used to prepare selectiveβ1-receptor antagonist atenolol and synthetic precursors of several non-ribosomal antibiotics[2].Moreover, p-hydroxymandelic acid shows promising applications in targeted drug delivery systems. But its market price is as high as $14,000 per kilogram, and its chemical synthesis methods have adverse environmental effects. Its biological methods have received a lot of attention recently.
The synthesis of HMA
As industries are evolving toward sustainable and environmental-friendly, microbial synthesis of these compounds attracted increasing attention[3]. Microbial metabolism has been harnessed to provide us access to natural products that are needed for the survival and reproduction of the host organisms, leading to the challenge in achieving high yields of desired products. In recent years, the iterative “design-build-test-learn” cycle of the synthetic biology has enabled rapid design and generation of highly diverse libraries. However, screening for desirable variants remains a critical bottleneck, underlining the need for high-throughput approaches to easily identify candidate phenotypes[4].

Inspiration
In recent years, the development of directed evolution has broadened the design scope of protein engineering, which can modify proteins without knowing the structural information and mechanism of action of target proteins. We boldly attempted directed evolution of strains, reasonably designed and altered metabolic pathways to obtain our target strains.
With the popularity of AlphaFold2 this year, we wondered if we could use protein structure prediction to better understand how individual amino acid site changes would affect the PobR protein pocket. So we tried to use bioinformatics methods to do molecular docking.
Approaches
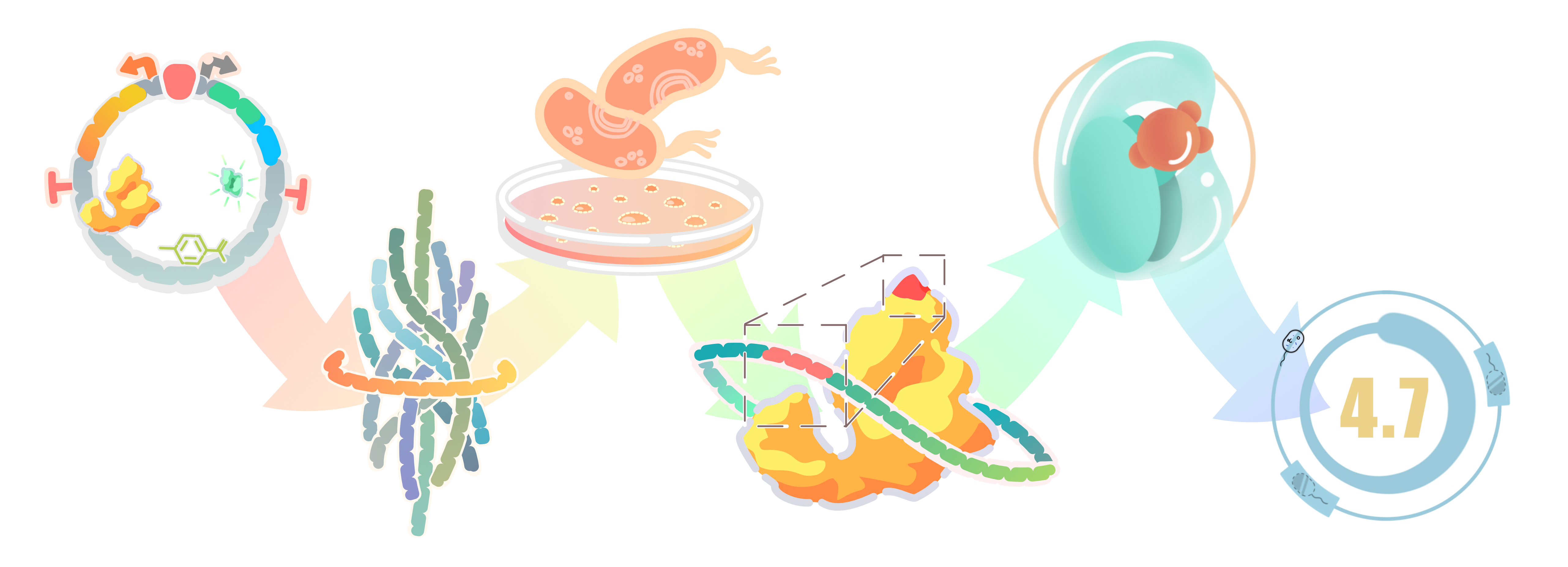
In this study, we aimed to develop an allosteric transcription factors (aTFs) based biosensor for HMA. There is no aTF that responds to HMA in nature, However, HMA has high structural similarity to 4HB in both core structure (a hydroxyphenyl) and side chains. PobR, an aTF for HMA analog 4-hydroxybenzoic acid, was used to alter its selectivity and create novel aTFs responsive to HMA by directed evolution.
We developed a library through random mutagenesis of PobR using error-prone PCR amplifications. Then we used high-throughput methods to screen out clones that respond to HMA, so we could used the HMA-responsive aTF improve the yield of HMA through directed evolution.
Our Procedure
References
- Reifenrath M and Boles E. Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae. Metab Eng, 2018, 45:246-254.
- Hubbard BK, Thomas MG and Walsh CT. Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem Biol, 2000, 7:931-942.
- Mitchler, M. M., Garcia, J. M., Montero, N. E., & Williams, G. J. . Transcription factor-based biosensors: a molecular-guided approach for natural product engineering. Curr Opin Biotechnol,2021,69:172-181.
- Cheng, F., Tang, X. L., & Kardashliev, T. Transcription Factor-Based Biosensors in High-Throughput Screening: Advances and Applications. Biotechnology Journal, 2018,13:7.