Results
Identifying the candidate residues of BmmGT1 for directed evolution
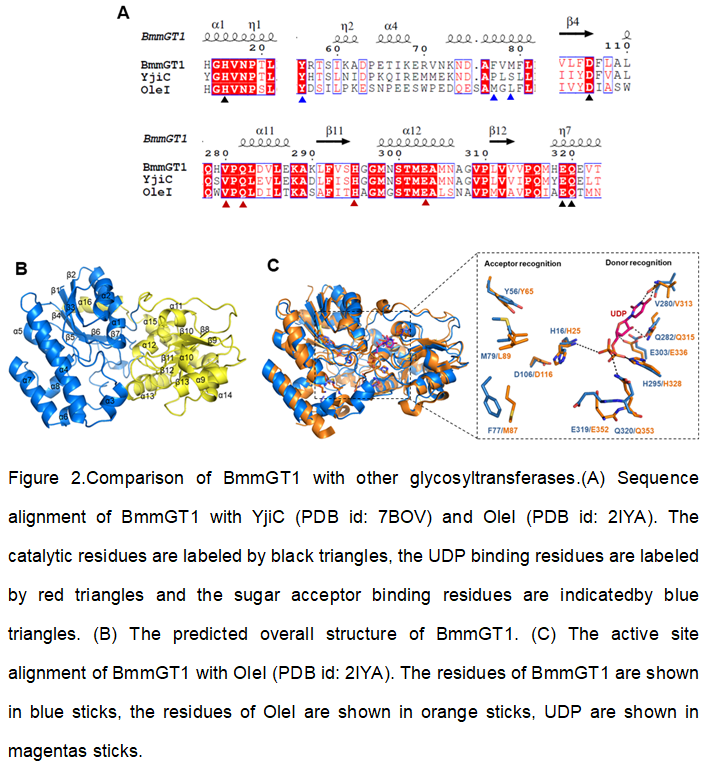
BmmGT1 shows 60% identity/78% similarity to the glycosyltransferase YjiC (WP_015252306) from Bacillussubtilis[26], 36% identity/54% similarity to OleI (ABA42118.2) from Streptomyces antibioticus[27]. Sequence alignment of these enzymes revealed thepresenceof the conserved catalytic residues His16, Asp106, Glu319 and Gln320 inBmmGT1 (Fig 2A). To find out the possible substrate binding residues in BmmGT1, we predicted its protein structure by using the online homology modeling program Phyre2[28]. BmmGT1 adopts a classic GT-B fold with two “Rossmann-like” β/α/βdomains(Fig 2B). The N-terminal domain (residues 4-206) is related to the recognition of sugar acceptors, and the C-terminal domain (residues 207-372) is responsible for the recognition of sugar donors (Fig 2B). The interface between the two domains houses the active site (Fig 2B). The binding pocket of BmmGT1 superimposed well with that of OleI[27] (PDB id: 2IYA, Fig 2C). Based on the substrate binding mode of OleI, the conserved residues Val280, Gln282, Glu303 and His295 in BmmGT1 are possibly responsible for the uridine diphosphate (UDP) binding, while Tyr56, Met79 and Phe77 might be related to the sugar acceptor binding (Fig 2C). Therefore, in order to improve the catalytic efficiency of BmmGT1 towards sugar acceptors, Tyr56, Met79 and Phe77 were selected for further directed evolution study.
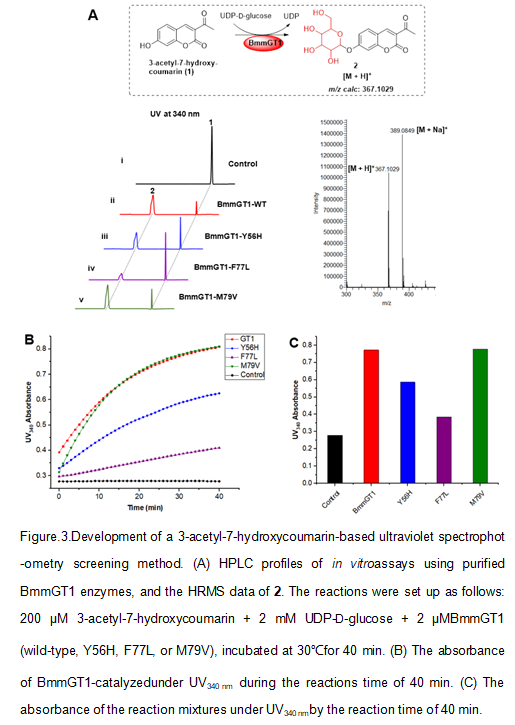
Development of a 3-acetyl-7-hydroxycoumarin-based ultraviolets pectrophotometry screening method
To develop an efficient screening method, we first checked if BmmGT1 is able to recognize 3-acetyl-7-hydroxycoumarin. As shown in Fig 3A, BmmGT1 indeed catalyzed glucosylation of 3-acetyl-7-hydroxycoumarin (panel i), which was confirmed by LC-HRMS. Considering the large π-π conjugated structure of 7-hydroxycoumarin, we detected the reaction by ultraviolet spectrophotometry and found that the attachment of a glycosylgroup at 7-OH of 3-acetyl-7-hydroxycoumarincouldsignificantly increase the UV absorbanceat 340 nm (Fig 3B). Therefore 3-acetyl-7-hydroxycoumarin-based ultraviolet spectrophotometric assaycould be exploited to detect the glycosylation activity of GTs. To evaluate the sensitivity and feasibility of the method, we constructed and purified the variants Y56H, F77L and M79V (Fig S2), followed by biochemical assay using 3-acetyl-7-hydroxycoumarin as sugar acceptor and UDP-D-glucose as sugar donor. The absorbance of the reaction mixtures was detected by spectrophotometer under UV340 nm per 1 min during the reaction time of 40 min. As shown in Fig 3B, with time going on, the absorbances of all the BmmGT1-catalyzed reactions increased gradually and almost reached plateau phase by 40 min.TheUV absorbance of M79V-catalyzed reaction is similar to that of the wild-type BmmGT1; the absorbances of Y56H- and F77L-catalyzed reaction are about three fourths and a half lower than that of the wild-type BmmGT1,respectively (Fig 3C). The parallel HPLC analysis result showed the conversion rates of these reactions were about 89.4% (wild-type; panel ii), 71.7% (Y56H; panel iii), 38.8% (F77L; panel iv) and 85.6% (M79V; panel v), which are consistent with those obtained by spectrophotometricassays.Therefore,an efficient 3-acetyl-7-hydroxycoumarin-based ultraviolet spectrophotometry screening method was developed for quick screening of the variants with enhanced glycosylation activities.
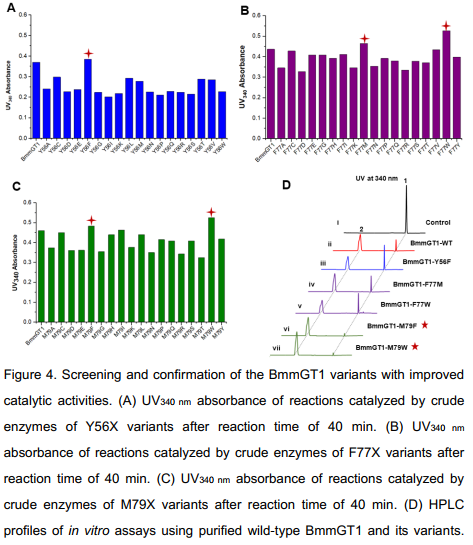
Saturation mutagenesis of sugar acceptor binding residues of BmmGT1
Given that Y56, F77, and M79 are probably involved in the binding of sugar acceptors (Fig 2C), they were selected for individual site saturation mutagenesis. Reverse complementary primers with mutation sites were designed to amplify the linear expression plasmids (pET28a/bmmGT1) with mutated base sequences (Table S2). Theresulting linear plasmids were ligated to circular molecules by seamless cloning (Fig S3). A total number of 57 expression vectors harboring site-mutated BmmGT1 at Y56, F77 and M79 were obtained, and were introduced into the expression host E. coli BL21 (DE3).
Screening of BmmGT1 variants with enhanced glycosylation activities
The expression strains were individually cultured, followed by addition of 0.2 mMIPTG to induce protein expression. Crude enzymes were subjected to screening by using the above established method. The crude enzymefromeachexpressionstrain was incubated with 3-acetyl-7-hydroxycoumarin and UDP-D-glucose in 96-well plates and the absorbance was measured under UV340 nm (Fig S4-S5).As shown in Fig 4A-4C, compared tothatof the wild-type BmmGT1, bythetimeof 40 min, the absorbance of Y56F increased by ~5.0% (Fig 4A); the absorbances of F77M and F77Wrespectivelyincreased by ~8.5% and 23.0% (Fig 4B); the absorbances of M79F and M79W respectively increased by ~6.6% and 17.5% (Fig4C). Thus, Y56F, F77M, F77W, M79F and M79W were identified as potential positive hits in the preliminary screening.
We thenpurified the screened mutants (Y56F, F77M, F77W, M79F and M79W), and measured their catalytic activities towards 3-acetyl-7-hydroxycoumarin and UDP-D-glucose by HPLC (Fig 4D and S6). As shown in Fig 4D, the catalytic activitiesof M79F (panel vi) and M79W (panel vii) increased by ~2.1-fold compared to that of the wild-type BmmGT1 (panel ii), while the other three mutants gave alittle bit lower conversion ratethan that of the wild-type BmmGT1. Therefore, through directed evolution, we successfully obtained the mutants M79F and M79W with enhanced glucosylation activity towards 3-acetyl-7-hydroxycoumarin.
The decreased catalytic activities of the purified variants Y56F, F77M and F77W led us to consider the differences of protein expression levels in the crude enzyme solution. Therefore, western blots were carried out to detect the expression of the wild-type BmmGT1 and its variants in the crude enzyme solution. However, the expression levels of Y56F, F77M and F77W are almost similar to that of the wild-type BmmGT1 (Fig S7). The increased UV absorbances of the reaction mixtures of Y56F, F77M and F77W crude enzyme might be due to unknown factors in cell extracts.
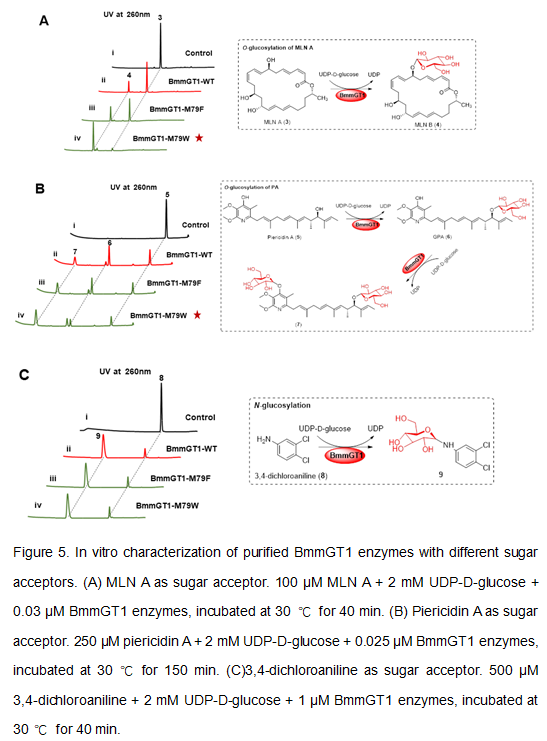
In vitro characterization of BmmGT1 variants with other sugar acceptors
Next, we tested the catalytic activities of M79F and M79Wtowardsthe 24-membered macrolide macrolactin A (MLN A), α-pyridone antibiotic piericidin A and 3,4-dichloroaniline. As shown in Fig 5 and S9, compared to the wild-type BmmGT1, the catalytic activities of M79W towards MLN A (Fig 5A, panel iv) and piericidin A (Fig 5B, panel iv) increased by ~3.6-fold and ~1.2-fold, respectively. While the catalytic activities of M79W towards 3,4-dichloroaniline (Fig 5C, panel iv), and the catalytic activities of M79F towards these three substrates (Fig 5A, panel iii; Fig 5B, panel iii; Fig 5C, panel iii) are similar to that of the wild-type BmmGT1 (Fig 5A, panel ii; Fig 5B, panel ii; Fig 5C, panel ii). These results indicated that M79W serves as an efficient enzyme for the O-glucosylation of MLN A and piericidin A as well, but M79F does not. These differences could be due to the binding mode disparity between different substrates in these mutants.
Discussion
Glycosylation is of crucial importance for the modification of natural products, improving their solubility, stability, and biological activity[1-3]. The Bacillus-derived glycosyltransferase BmmGT1 exhibited broad substrate promiscuity but with low catalytic efficiency[18-20]. In an effort to enhance the glycosylation activities of Bacillus-derived GT BmmGT1 towards different sugar acceptors, we performed saturation mutations of the sugar acceptor binding sites.Using a 3-acetyl-7-hydroxycoumarin-based ultraviolet spectrophotometry screening method, weobtained two variants, M79F and M79W, withenhanced catalytic activities by ~2.1-fold. Of note, the glucosylation activities of M79W towards MLN A and piericidin A increased by ~3.6-fold and ~1.2-fold, respectively.
Coumarins is one of the most common skeletons of natural products and are widely distributed in many plant species[21]. Naturally occurring coumarins often exist as glycosides, and these derivatives exhibited a broad spectrum of pharmacological activities, including antioxidant[29], anti-cancer[30], anti-inflammatory[31] etc. So far, the reports about GTs being able to transfer the sugar moiety onto coumarins are limited and the catalytic efficiencies are not high[31-33].In our study, a Bacillus-derived GT BmmGT1 was found to be able to catalyze the glucosylation of 3-acetyl-7-hydroxycoumarin. Accordingly, an efficient screening method wasestablished.By rational design and directed evolution, M79F and M79W with enhanced catalytic efficiencies towards 3-acetyl-7-hydroxycoumarin, were identified. Detection of M79F and M79W with alternative coumarin derivatives are going on. These results make the enzyme an effective tool in the enzymatic modification to generate bioactive coumarin glycosides.
The wild-type BmmGT1 exhibited broad substrate flexibility towards the sugar acceptors[18-20]. The BmmGT1 variants M79F and M79W also recognized all the substates tested (Fig 4D and Fig 5). Compared to the wild-type BmmGT1, the activities of M79W towards MLN A and piericidin A alsoincreased by ~3.6-foldand 1.2-fold, respectively. As theyall have cyclic structures (24-membered macro-ring or 6-membered ring), substitution of Met79 to Trp could generate strong π-π interactions, facilitating substrates binding. The disparity of the binding mode with different substrates could lead to catalytic efficiency differences of the enzymes towards various substrates. More substrates need to be tested to probe the substrate flexibility of the BmmGT1 variants. The residues predicted to have hydrophobic interactions with sugar acceptors could serve as potential effective residues for future GT engineering.
In conclusion, using 3-acetyl-7-hydroxycoumarin-based ultraviolet spectrophotometry screening method, BmmGT1 variants with improved catalytic activity were successfully obtained via directed evolution. Given the substrate promiscuity, the BmmGT1 variants could be exploited as valuable biocatalysts toglycodiversify small molecules in the search for drug candidates.